Hi.
I’ve recently encountered the fact that the covariance matrix can be scaled or unscaled when a minimization is performed using the Levenberg Marquardt Method lmfit.minimize(…). How is the scaling performed?
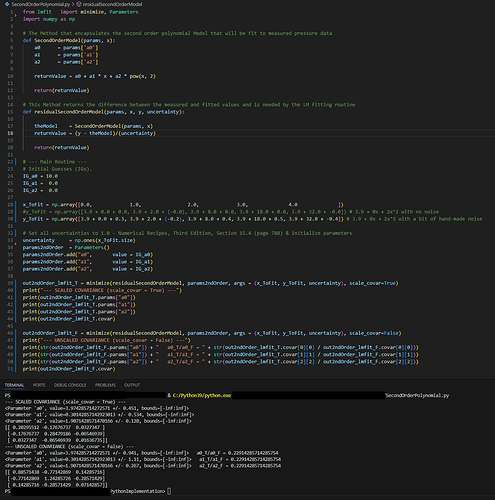
An example of this is found in the code below which fits a second order polynomial of the form y = a0 + a1x + a2x^2 to 5 points. The covariance matrix for the fit is different depending on whether the scale_covar pamameter is set to True (T) or False (F). However, the ratio of the diagonal elements is the same for elements [0][0], [1][1] and [2][2] (i.e., 0.22914285… in the attached screenshot). How is this scaling factor calcluated?
Thanks,
Eric
from lmfit import minimize, Parameters
import numpy as np
# The Method that encapsulates the second order polynomial Model that will be fit to measured pressure data
def SecondOrderModel(params, x):
a0 = params['a0']
a1 = params['a1']
a2 = params['a2']
returnValue = a0 + a1 * x + a2 * pow(x, 2)
return(returnValue)
# This Method returns the difference between the measured and fitted values and is needed by the LM fitting routine
def residualSecondOrderModel(params, x, y, uncertainty):
theModel = SecondOrderModel(params, x)
returnValue = (y - theModel)/(uncertainty)
return(returnValue)
# --- Main Routine ---
# Initial Guesses (IGs).
IG_a0 = 10.0
IG_a1 = 0.0
IG_a2 = 0.0
x_ToFit = np.array([0.0, 1.0, 2.0, 3.0, 4.0 ])
#y_ToFit = np.array([3.9 + 0.0 + 0.0, 3.9 + 2.0 + (-0.0), 3.9 + 8.0 + 0.0, 3.9 + 18.0 + 0.0, 3.9 + 32.0 + -0.0]) # 3.9 + 0x + 2x^2 with no noise
y_ToFit = np.array([3.9 + 0.0 + 0.3, 3.9 + 2.0 + (-0.2), 3.9 + 8.0 + 0.4, 3.9 + 18.0 + 0.5, 3.9 + 32.0 + -0.4]) # 3.9 + 0x + 2x^2 with a bit of hand-made noise
# Set all uncertainties to 1.0 - Numerical Recipes, Third Edition, Section 15.4 (page 788) & initialize parameters
uncertainty = np.ones(x_ToFit.size)
params2ndOrder = Parameters()
params2ndOrder.add("a0", value = IG_a0)
params2ndOrder.add("a1", value = IG_a1)
params2ndOrder.add("a2", value = IG_a2)
out2ndOrder_lmfit_T = minimize(residualSecondOrderModel, params2ndOrder, args = (x_ToFit, y_ToFit, uncertainty), scale_covar=True)
print("--- SCALED COVARIANCE (scale_covar = True) ---")
print(out2ndOrder_lmfit_T.params["a0"])
print(out2ndOrder_lmfit_T.params["a1"])
print(out2ndOrder_lmfit_T.params["a2"])
print(out2ndOrder_lmfit_T.covar)
out2ndOrder_lmfit_F = minimize(residualSecondOrderModel, params2ndOrder, args = (x_ToFit, y_ToFit, uncertainty), scale_covar=False)
print("--- UNSCALED COVARIANCE (scale_covar = False) ---")
print(str(out2ndOrder_lmfit_F.params["a0"]) + " a0_T/a0_F = " + str(out2ndOrder_lmfit_T.covar[0][0] / out2ndOrder_lmfit_F.covar[0][0]))
print(str(out2ndOrder_lmfit_F.params["a1"]) + " a1_T/a1_F = " + str(out2ndOrder_lmfit_T.covar[1][1] / out2ndOrder_lmfit_F.covar[1][1]))
print(str(out2ndOrder_lmfit_F.params["a2"]) + " a2_T/a2_F = " + str(out2ndOrder_lmfit_T.covar[2][2] / out2ndOrder_lmfit_F.covar[2][2]))
print(out2ndOrder_lmfit_F.covar)